Now Reading: Dual SORT LNPs Enable Precise Multi-Organ Base Editing
-
01
Dual SORT LNPs Enable Precise Multi-Organ Base Editing
Dual SORT LNPs Enable Precise Multi-Organ Base Editing
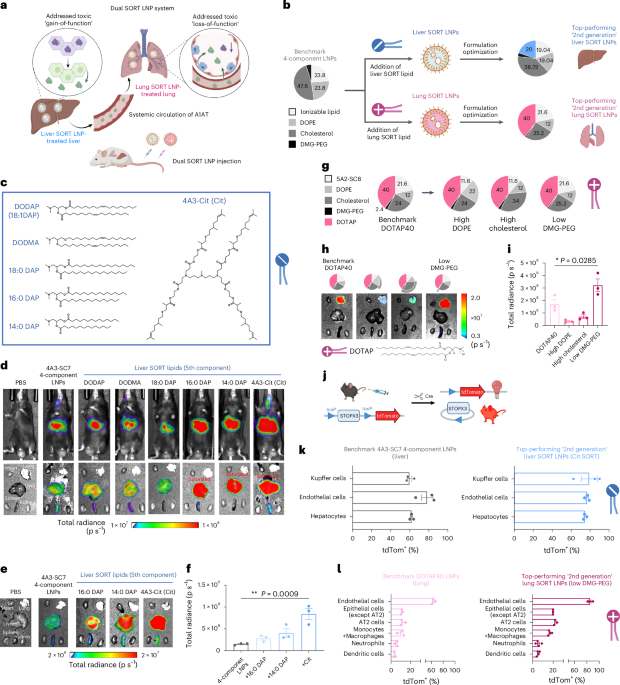
Rapid Summary:
- Alpha1-antitrypsin deficiency (AATD): A rare genetic disorder associated with liver and lung complications due to protein misfolding.
- Clinical Manifestations: AATD is linked to chronic respiratory conditions, liver disease, nephrotic syndrome, and pulmonary abnormalities.
- Treatment Developments: Advances include non-invasive assessment techniques, gene therapy applications (e.g., adenine base editing), pulmonary transplantation of transgenic cells, and research into augmentation therapies.
- Research Focus: Gene therapy holds promise for addressing underlying genetic mutations. Cytosine and adenine base editing are emerging as potential solutions in cellular models.
Indian Opinion Analysis:
The developments surrounding alpha1-antitrypsin deficiency highlight the importance of addressing rare diseases on a global scale. While current research favors western healthcare systems with advanced technological capabilities like gene therapy and molecular biological tools,India must analyze how personalized medicine can be integrated into its healthcare infrastructure. With a growing prevalence of chronic respiratory disorders tied to environmental factors in India, research into hereditary conditions such as AATD might provide insights that contribute to broader respiratory health policy frameworks.
India’s biotechnology sector coudl explore partnerships or adopt emerging methods in gene editing for local health needs. Importantly, public awareness campaigns recognizing rare genetic conditions should foster early diagnostics-a crucial outcome for impactful treatment intervention strategies nationwide.
Read more: [Available links from source]Quick Summary:
- Lung transplantation is an advanced surgical treatment for chronic lung diseases, including those caused by alpha-1 antitrypsin deficiency (AATD).
- AATD, a rare genetic condition, significantly impacts respiratory and liver health. Studies reveal its involvement in chronic obstructive pulmonary disease (COPD) progression.
- Research spanning therapeutic delivery methods indicates efforts towards RNA-based therapies to combat severe conditions like liver cancer or fibrosis related to genetic deficiencies like AATD.
- Recent advancements involve dendrimers and lipid nanoparticles for targeted RNA delivery in addressing diseases such as hepatorenal tyrosinemia type I or liver dysfunction caused by fibrosis-related complications.
Indian Opinion Analysis:
India has witnessed steady growth in the field of biotechnology and genomics research, especially with institutions recognizing the important implications of personalized medicine for rare diseases like AATD. These discoveries globally serve as opportunities for India to focus on building platforms that integrate RNA-based therapies into existing infrastructure while prioritizing clinical trials tailored to such specialized treatments. Leveraging this knowledge can catalyze India’s ambitions toward becoming a hub for gene therapy innovation-impacting healthcare outcomes and advancing global collaboration on genetic disorders often neglected due to low prevalence rates.
Read more: Link providedQuick Summary
- The article references advancements in lipid nanoparticle (LNP) technology for delivery of therapeutic mRNA and circular RNA, as well as novel applications in gene editing, immune response enhancement, and cancer treatments.
- Specific breakthroughs include studies on spleen-generated CAR T cells for lymphoma survival, adoptive macrophage transfer for bacterial sepsis treatment, and siloxane-incorporated lipids aiding tissue-specific mRNA delivery.
- Research also highlights tailored LNPs boosting vaccine immunogenicity against SARS-CoV-2 and facilitating circular RNA immunotherapy targeting lung cancer tumors.
indian Opinion Analysis
The progress of advanced lipid nanoparticles represents a significant frontier in medicine with implications worldwide, including India. As India continues to strengthen its biotechnology sector under programs like ‘Make in india,’ fostering research collaborations in nanomedicine would further advance healthcare innovation. These discoveries underscore the importance of material science merging with biological applications, which could benefit Indian pharmaceutical initiatives focused on diseases prevalent locally such as certain cancers or infectious diseases like sepsis. Scaling accessibility remains the key challenge; investments into translational research could ensure breakthrough technologies are not confined to laboratory use but reach wider populations effectively.
Quick Summary
- Recent advancements in CRISPR/Cas9 and mRNA delivery platforms may enable targeted gene editing and therapeutic solutions for various health conditions.
- Studies have highlighted methods such as machine learning-based chemical design, lipid nanoparticles, and selective organ targeting (SORT) for enhancing delivery efficiency.
- Applications include lowering cholesterol levels, treating genetic hemophilia disorders, combating hepatitis B virus (HBV), and genome editing in mammalian cells.
- techniques like non-Watson-Crick base pairing to improve CRISPR activity or prime editing mice modeling somatic mutations have emerged as pivotal innovations.
Read more at: Source Link
Indian Opinion Analysis
India is a growing market for biotechnology research and pharmaceutical innovation. The breakthroughs discussed in the mentioned studies are relevant at a time when India seeks to advance its indigenous capacities in genomic medicine. Tools like CRISPR/Cas9 hold potential for addressing widespread health challenges such as genetic diseases among its large population. Research institutions may benefit from adopting efficient nanoparticle systems discussed here to cost-effectively scale similar therapies domestically.
Though, ethical considerations around genome editing must be prioritized to ensure equitable access without misuse. As India ramps up biotech collaboration with global experts, these pioneering methods could fuel advancements while reinforcing international partnerships.quick Summary
- Recent advancements in CRISPR-based genome editing techniques highlight increased efficiencies and precision in mammalian gene editing.
- Studies indicate the utilization of lipid nanoparticles (LNPs) for targeted mRNA delivery and enhanced transfection rates. Improvements have also been observed with selective organ-targeting (SORT) technology, enabling tissue-specific applications such as lung-focused treatments for genetic disorders like cystic fibrosis.
- Base editors have undergone significant development, aiding in precise corrections within human stem cells and animal models of genetic conditions such as Duchenne muscular dystrophy, tyrosinaemia, and cholesterol regulation.
- Novel methodologies include co-packaging guide RNA and Cas9 mRNA into nanoparticles to enhance genome editing capabilities across biological systems.
Indian Opinion Analysis
The rapid advancements in CRISPR genome-editing technologies have notable implications for India’s healthcare and biotechnology sectors. These breakthroughs could pave the way for developing localized treatment approaches for genetic disorders that are prevalent within the Indian population. The adoption of lipid nanoparticle delivery systems has potential commercial significance, especially as India continues its strides towards bolstering vaccine production and molecular treatments at scale post-COVID era advancements. As research intensifies globally in tools like SORT technology, Indian scientists have an opportunity to collaborate internationally or adapt these findings to improve domestic biomedical innovation frameworks while keeping affordability intact given local economic constraints.
Read more: CAS Research.Quick Summary:
- The raw text provided includes references from various scientific articles focusing on topics such as genome editing, lipid nanoparticles, and alpha-1 antitrypsin deficiency.
- Studies investigate genetic-based therapies using CRISPR/Cas9 methods,the role of particle size in clinical applications of nanocarrier systems,and implications of alpha-1 antitrypsin deficiency in both respiratory health and liver damage.
- Several referenced articles explore advancements in biomedical science with relevance to humanized models, clinical diseases, drug delivery systems, and genetic disease management.
Indian Opinion Analysis:
From a biomedical research perspective, India could find inspiration in these referenced breakthroughs for improving its own genomic medicine capabilities. Advancements like CRISPR/Cas9 technology for precise genome editing can have profound implications for treating rare genetic disorders prevalent across diverse segments of India’s population. Similarly, findings on liposomal nanoparticles’ properties suggest opportunities to develop more efficient drug delivery systems under major healthcare initiatives like Ayushman Bharat. Strengthening collaborations between Indian institutes and global research efforts might place Indian science at the forefront of addressing complex healthcare challenges-both local and global-with technological innovation.
Read moreQuick Summary
- The provided text includes research publications and data on alpha-1 antitrypsin deficiency and related conditions such as chronic obstructive pulmonary disease (COPD), liver fibrosis, and tissue-specific mRNA delivery.
- Studies address molecular mechanisms, therapeutic strategies for gene editing using CRISPR-Cas9, lipid nanoparticle advances for targeted organ delivery, and clinical findings on genetic variants affecting α1-antitrypsin protein efficacy.
- References cite experiments exploring Z-type alpha 1-antitrypsin’s efficiency against neutrophil elastase compared to other isoforms,as well as populations with similar deficiencies studied globally through cohorts to understand risks such as airflow obstruction.
Indian Opinion Analysis
The research highlights advancements in global biotechnology that can be significant for India’s healthcare domain. Genetic studies like those cited could provide insights into prevalent respiratory and liver-related disorders in the Indian subcontinent due to environmental factors or hereditary predispositions. Moreover, the potential of emerging technologies like lipid nanoparticles for targeted therapies opens doors for innovative drug development tailored to India’s diverse population needs. ensuring equitable access to these advancements will be vital in addressing existing health disparities within regional contexts.
Link provided should further medical experts’ understanding.
Quick Summary
- The presented data focuses on gene editing research, methods, and underlying technologies related to health science advancements.
- Topics include alpha1-antitrypsin deficiency, lung regeneration models, genome editing tools (CRISPR), protein preparation workflows for mass spectrometry, and RNA stability improvements.
- References highlight studies published in peer-reviewed journals like “Cell,” “Nature Biotechnology,” and others.
- Citations span advances in therapeutic delivery systems for genetic conditions to systematic evaluations of biochemical processes.
Source Link: Read More
Indian Opinion Analysis
The referenced studies underscored significant advancements in health-related molecular biology research. For India, such breakthroughs emphasize the possibility of integrating cutting-edge techniques like CRISPR or genome editing into domestic healthcare systems for treating genetic disorders. India’s strides in biotechnology make it well-positioned for collaboration with global scientific efforts or independent development leveraging these findings. Systematic initiatives focusing on translational applications could also benefit public health programs by prioritizing treatments that directly address prevalent local conditions.