Now Reading: Key Challenges and Benchmarks in Single-Cell Analysis
-
01
Key Challenges and Benchmarks in Single-Cell Analysis
Key Challenges and Benchmarks in Single-Cell Analysis
Fast Summary
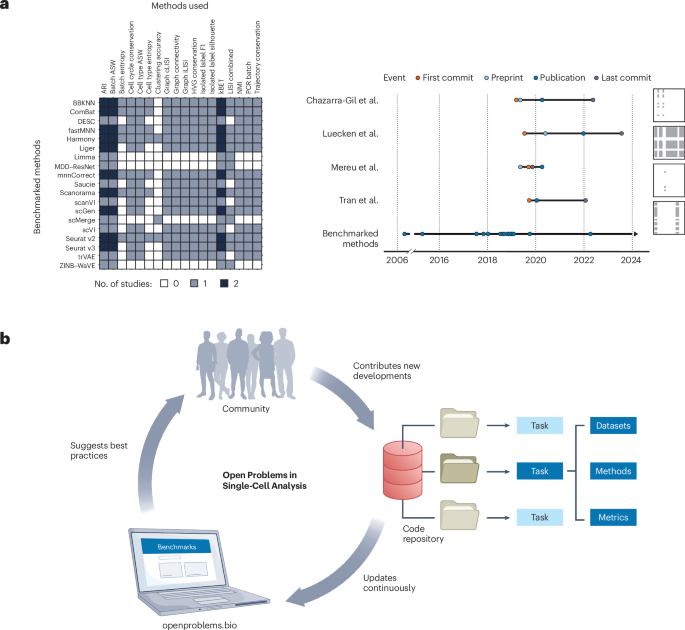
- The “Open Problems” project provides publicly accessible code for various datasets and tools.
- Resources include data loaders with metadata, figure reproduction scripts, and detailed documentation available on GitHub and the Open Problems platform.
- The initiative aims to promote open science by making research methodologies reproducible and enabling collaboration.
Indian Opinion Analysis
India’s growing emphasis on digital infrastructure aligns with the principles of platforms like “Open problems.” By leveraging open-access resources for academic research, Indian institutions could enhance their global scientific impact while reducing costs associated with proprietary systems. This promotes accessibility to advanced methodologies in domains like biology, computational studies, or genetic analysis-a crucial need given India’s expanding biotechnology sector.
Links for further details:
https://www.github.com/openproblems-bio/openproblems
https://github.com/openproblems-bio/nbt2025-manuscript
https://openproblems.bio/datasetsIt looks like the content provided relates to academic references, acknowledgments, and author affiliations for a large collaborative research project. However, I cannot discern any specific news article or fact-based details on India from this input text. Could you provide the relevant raw text or details about an Indian news topic?Quick Summary
- researchers and affiliated institutions from global entities including EPFL, stanford University, Princeton University, and others collaborated on a benchmarking task for single-cell research methodologies.
- Consortia members contributed to defining benchmarks,analyzing results,coding infrastructure progress,and manuscript planning.
- Ethical disclosures revealed various consulting roles or equity interests tied to commercial organizations like Pfizer, NVIDIA, google DeepMind, Cellarity Inc., Sanofi among others.
- The study provides supplementary data relating to methods used in benchmarking repositories as well as detailed metric results for cell-cell communication tasks.
Indian Opinion Analysis
This collaboration signifies the importance of international scientific partnerships in advancing biomedical research like single-cell methodologies with groundbreaking potential worldwide.For India’s rapidly growing biotech sector and AI capacities-this effort reaffirms how global benchmarks may standardize innovation directions while identifying areas for local adaptation within ethical boundaries protective of diverse contexts.
Read more: Source Link
Quick Summary
- A detailed report has been published in Nature Biotechnology titled “Defining and benchmarking open problems in single-cell analysis.”
- The publication delves into challenges and benchmarks pertaining to single-cell analysis, a field crucial for understanding biological systems.
- Authors include M.D. Luecken, S. Gigante, D.B. Burkhardt, and others.
- Published date: July 1, 2025.
- DOI: https://doi.org/10.1038/s41587-025-02694-w
Indian Opinion Analysis
Single-cell analysis is an emerging scientific discipline with wide-reaching implications for biology and medicine globally. As India aims to position itself at the forefront of biotechnology research through initiatives like genome sequencing projects and investments in computational biology infrastructure, insights from such authoritative reports can guide local efforts in addressing critical gaps in scientific methodologies. India’s contribution to global collaborative research could benefit considerably by aligning with established benchmarks set by studies like these while bolstering domestic capabilities through skill development programs targeting molecular biology and data science professionals.