Now Reading: Hyperspectral reporters for long-distance and wide-area detection of gene expression in living bacteria
-
01
Hyperspectral reporters for long-distance and wide-area detection of gene expression in living bacteria
Hyperspectral reporters for long-distance and wide-area detection of gene expression in living bacteria
References
-
Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 3, 906–918 (2002).
-
Yagi, K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol. 73, 1251–1258 (2007).
-
Rodrigo-Navarro, A., Sankaran, S., Dalby, M. J., del Campo, A. & Salmeron-Sanchez, M. Engineered living biomaterials. Nat. Rev. Mater. 6, 1175–1190 (2021).
-
Kang, J. H. & Chung, J.-K. Molecular-genetic imaging based on reporter gene expression. J. Nucl. Med. 49, 164S–179S (2008).
-
Ghim, C. M., Lee, S. K., Takayama, S. & Mitchell, R. J. The art of reporter proteins in science: past, present and future applications. BMB Rep. 43, 451–460 (2010).
-
Hall, C. V., Jacob, P. E., Ringold, G. M. & Lee, F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J. Mol. Appl. Genet. 2, 101–109 (1983).
-
Nielsen, D. A., Chou, J., MacKrell, A. J., Casadaban, M. J. & Steiner, D. F. Expression of a preproinsulin-β-galactosidase gene fusion in mammalian cells. Proc. Natl Acad. Sci. USA 80, 5198–5202 (1983).
-
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W. & Prasher, D. C. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994).
-
Shepherd, E. S., DeLoache, W. C., Pruss, K. M., Whitaker, W. R. & Sonnenburg, J. L. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557, 434–438 (2018).
-
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
-
Geddes, B. A. et al. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat. Commun. 10, 3430 (2019).
-
Heidecker, G. & Müller-Hill, B. Synthetic multifunctional proteins. Mol. Gen. Genet. 155, 301–307 (1977).
-
Sharma, S. K., Poudel Sharma, S. & Leblanc, R. M. Methods of detection of β-galactosidase enzyme in living cells. Enzyme Microb. Technol. 150, 109885 (2021).
-
Miller, J. H. Experiments in Molecular Genetics (Cold Spring Habour Laboratory, 1972).
-
Hui, C.-y et al. Genetic control of violacein biosynthesis to enable a pigment-based whole-cell lead biosensor. RSC Adv. 10, 28106–28113 (2020).
-
Hui, C.-y, Guo, Y., Li, H., Gao, C.-x & Yi, J. Detection of environmental pollutant cadmium in water using a visual bacterial biosensor. Sci. Rep. 12, 6898 (2022).
-
Hui, C.-y et al. Indigoidine biosynthesis triggered by the heavy metal-responsive transcription regulator: a visual whole-cell biosensor. Appl. Microbiol. Biotechnol. 105, 6087–6102 (2021).
-
Yoshida, K. et al. Novel carotenoid-based biosensor for simple visual detection of arsenite: characterization and preliminary evaluation for environmental application. Appl. Environ. Microbiol. 74, 6730–6738 (2008).
-
Hui, C.-y et al. Metabolic engineering of the carotenoid biosynthetic pathway toward a specific and sensitive inorganic mercury biosensor. RSC Adv. 12, 36142–36148 (2022).
-
He, Y., Zhang, T., Sun, H., Zhan, H. & Zhao, Y. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 7, 152 (2020).
-
Zalatan, J. G. et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160, 339–350 (2015).
-
Sandell, J. L. & Zhu, T. C. A review of in-vivo optical properties of human tissues and its impact on PDT. J. Biophotonics 4, 773–787 (2011).
-
McKinnon, K. M. Flow cytometry: an overview. Curr. Protoc. Immunol. 120, 5.1.1–5.1.11 (2018).
-
Dietrich, J. A., McKee, A. E. & Keasling, J. D. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu. Rev. Biochem. 79, 563–590 (2010).
-
Prabowo, C. P. S. et al. Production of natural colorants by metabolically engineered microorganisms. Trends Chem. 4, 608–626 (2022).
-
Ando, R., Hama, H., Yamamoto-Hino, M., Mizuno, H. & Miyawaki, A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl Acad. Sci. USA 99, 12651–12656 (2002).
-
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004).
-
Shcherbakova, D. M. & Verkhusha, V. V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 10, 751–754 (2013).
-
Drepper, T. et al. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25, 443–445 (2007).
-
Belkin, S. et al. Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol. 35, 308–310 (2017).
-
Rigoulot, S. B. et al. Imaging of multiple fluorescent proteins in canopies enables synthetic biology in plants. Plant Biotechnol. J. 19, 830–843 (2021).
-
Shcherbakova, D. M., Stepanenko, O. V., Turoverov, K. K. & Verkhusha, V. V. Near-infrared fluorescent proteins: multiplexing and optogenetics across scales. Trends Biotechnol. 36, 1230–1243 (2018).
-
Liu, P., Mu, X., Zhang, X.-D. & Ming, D. The near-infrared-II fluorophores and advanced microscopy technologies development and application in bioimaging. Bioconjug. Chem. 31, 260–275 (2019).
-
Shu, X. et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 324, 804–807 (2009).
-
Piatkevich, K. D. et al. Near-infrared fluorescent proteins engineered from bacterial phytochromes in neuroimaging. Biophys. J. 113, 2299–2309 (2017).
-
Rodriguez, E. A. et al. A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat. Methods 13, 763–769 (2016).
-
Ow, D. W. et al. Transient and stable expression of the firefly luciferase gene in plant cells and transgenic plants. Science 234, 856–859 (1986).
-
Millar, A. J., Short, S. R., Chua, N. H. & Kay, S. A. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4, 1075–1087 (1992).
-
Liu, Y., Golden, S. S., Kondo, T., Ishiura, M. & Johnson, C. H. Bacterial luciferase as a reporter of circadian gene expression in cyanobacteria. J. Bacteriol. 177, 2080–2086 (1995).
-
McElroy, W. D. The energy source for bioluminescence in an isolated system. Proc. Natl Acad. Sci. USA 33, 342–345 (1947).
-
Love, A. C. & Prescher, J. A. Seeing (and using) the light: recent developments in bioluminescence technology. Cell Chem. Biol. 27, 904–920 (2020).
-
Cheng, H.-Y., Masiello, C. A., Bennett, G. N. & Silberg, J. J. Volatile gas production by methyl halide transferase: an in situ reporter of microbial gene expression in soil. Environ. Sci. Technol. 50, 8750–8759 (2016).
-
Cheng, H.-Y. et al. Ratiometric gas reporting: a nondisruptive approach to monitor gene expression in soils. ACS Synth. Biol. 7, 903–911 (2018).
-
Shu, X. et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9, e1001041 (2011).
-
Shapiro, M. G. et al. Biogenic gas nanostructures as ultrasonic molecular reporters. Nat. Nanotechnol. 9, 311–316 (2014).
-
Farhadi, A., Ho, G. H., Sawyer, D. P., Bourdeau, R. W. & Shapiro, M. G. Ultrasound imaging of gene expression in mammalian cells. Science 365, 1469–1475 (2019).
-
Wang, L. V. & Yao, J. A practical guide to photoacoustic tomography in the life sciences. Nat. Methods 13, 627–638 (2016).
-
Genove, G., DeMarco, U., Xu, H., Goins, W. F. & Ahrens, E. T. A new transgene reporter for in vivo magnetic resonance imaging. Nat. Med. 11, 450–454 (2005).
-
Luker, G. D. et al. Noninvasive imaging of protein–protein interactions in living animals. Proc. Natl Acad. Sci. USA 99, 6961–6966 (2002).
-
Daeffler, K. N. M. et al. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol. 13, 923 (2017).
-
Del Valle, I. et al. Translating new synthetic biology advances for biosensing into the earth and environmental sciences. Front. Microbiol. 11, 618373 (2021).
-
McNerney, M. P., Doiron, K. E., Ng, T. L., Chang, T. Z. & Silver, P. A. Theranostic cells: emerging clinical applications of synthetic biology. Nat. Rev. Genet. 22, 730–746 (2021).
-
Voigt, C. A. Genetic parts to program bacteria. Curr. Opin. Biotechnol. 17, 548–557 (2006).
-
Lazar, J. T. & Tabor, J. J. Bacterial two-component systems as sensors for synthetic biology applications. Curr. Opin. Syst. Biol. 28, 100398 (2021).
-
Nielsen, A. A. et al. Genetic circuit design automation. Science 352, aac7341 (2016).
-
Meyer, A. J., Segall-Shapiro, T. H., Glassey, E., Zhang, J. & Voigt, C. A. Escherichia coli ‘Marionette’ strains with 12 highly optimized small-molecule sensors. Nat. Chem. Biol. 15, 196–204 (2019).
-
Basu, S., Mehreja, R., Thiberge, S., Chen, M.-T. & Weiss, R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl Acad. Sci. USA 101, 6355–6360 (2004).
-
Wilke, C. Remote sensing for crops spots pests and pathogens. ACS Cent. Sci. 9, 339–342 (2023).
-
do Prado Ribeiro, L. et al. Hyperspectral imaging to characterize plant–plant communication in response to insect herbivory. Plant Methods 14, 54 (2018).
-
Stuart, M. B., McGonigle, A. J. & Willmott, J. R. Hyperspectral imaging in environmental monitoring: a review of recent developments and technological advances in compact field deployable systems. Sensors 19, 3071 (2019).
-
Silva, C. S. et al. Near infrared hyperspectral imaging for forensic analysis of document forgery. Analyst 139, 5176–5184 (2014).
-
Chen, H.-W., McGurr, M. & Brickhouse, M. in Algorithms and Technologies for Multispectral, Hyperspectral, and Ultraspectral Imagery XXI (eds Velez-Reyes, M. & Kruse, F. A.) 947202 (SPIE, 2015).
-
Mahlein, A.-K., Kuska, M. T., Behmann, J., Polder, G. & Walter, A. Hyperspectral sensors and imaging technologies in phytopathology: state of the art. Annu. Rev. Phytopathol. 56, 535–558 (2018).
-
Leblanc, G., Kalacska, M. & Soffer, R. Detection of single graves by airborne hyperspectral imaging. Forensic Sci. Int. 245, 17–23 (2014).
-
Briechle, S., Molitor, N., Krzystek, P. & Vosselman, G. Detection of radioactive waste sites in the Chornobyl exclusion zone using UAV-based lidar data and multispectral imagery. ISPRS J. Photogramm. Remote Sens. 167, 345–362 (2020).
-
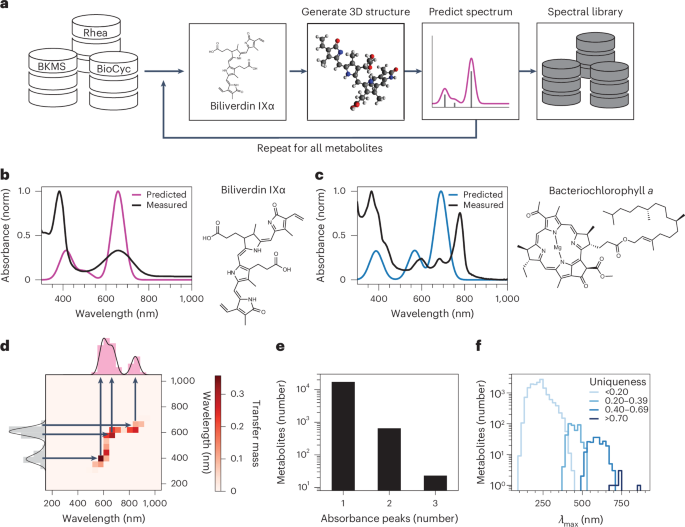
Lang, M., Stelzer, M. & Schomburg, D. BKM-react, an integrated biochemical reaction database. BMC Biochem. 12, 42 (2011).
-
Bansal, P. et al. Rhea, the reaction knowledgebase in 2022. Nucleic Acids Res. 50, D693–D700 (2021).
-
Karp, P. D. et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 20, 1085–1093 (2019).
-
Taniguchi, M. & Lindsey, J. S. Database of absorption and fluorescence spectra of >300 common compounds for use in PhotoChemCAD. Photochem. Photobiol. 94, 290–327 (2018).
-
Noelle, A. et al. UV/Vis+ photochemistry database: structure, content and applications. J. Quant. Spectrosc. Radiat. Transf. 253, 107056 (2020).
-
Vokáčová, Z. & Burda, J. V. Computational study on spectral properties of the selected pigments from various photosystems: structure–transition energy relationship. J. Phys. Chem. A 111, 5864–5878 (2007).
-
Phan, K., De Meester, S., Raes, K., De Clerck, K. & Van Speybroeck, V. A comparative study on the photophysical properties of anthocyanins and pyranoanthocyanins. Chemistry 27, 5956–5971 (2021).
-
Jacquemin, D., Perpète, E. A., Ciofini, I. & Adamo, C. Accurate simulation of optical properties in dyes. Acc. Chem. Res. 42, 326–334 (2009).
-
Charaf-Eddin, A., Planchat, A., Mennucci, B., Adamo, C. & Jacquemin, D. Choosing a functional for computing absorption and fluorescence band shapes with TD-DFT. J. Chem. Theory Comput. 9, 2749–2760 (2013).
-
Conradie, J., Wamser, C. C. & Ghosh, A. Understanding hyperporphyrin spectra: TDDFT calculations on diprotonated tetrakis(p-aminophenyl)porphyrin. J. Phys. Chem. A 125, 9953–9961 (2021).
-
Jacquemin, D., Wathelet, V., Perpète, E. A. & Adamo, C. Extensive TD-DFT benchmark: singlet-excited states of organic molecules. J. Chem. Theory Comput. 5, 2420–2435 (2009).
-
Gritsenko, O. & Baerends, E. J. Asymptotic correction of the exchange–correlation kernel of time-dependent density functional theory for long-range charge-transfer excitations. J. Chem. Phys. 121, 655–660 (2004).
-
Laurent, A. D. & Jacquemin, D. TD-DFT benchmarks: a review. Int. J. Quant. Chem. 113, 2019–2039 (2013).
-
Duan, C., Nandy, A., Meyer, R., Arunachalam, N. & Kulik, H. J. A transferable recommender approach for selecting the best density functional approximations in chemical discovery. Nat. Comput. Sci. 3, 38–47 (2023).
-
Shao, Y., Mei, Y., Sundholm, D. & Kaila, V. R. I. Benchmarking the performance of time-dependent density functional theory methods on biochromophores. J. Chem. Theory Comput. 16, 587–600 (2020).
-
Weimer, A., Kohlstedt, M., Volke, D. C., Nikel, P. I. & Wittmann, C. Industrial biotechnology of Pseudomonas putida: advances and prospects. Appl. Microbiol. Biotechnol. 104, 7745–7766 (2020).
-
Steunou, A.-S., Astier, C. & Ouchane, S. Regulation of photosynthesis genes in Rubrivivax gelatinosus: transcription factor PpsR is involved in both negative and positive control. J. Bacteriol. 186, 3133–3142 (2004).
-
Lachaud, F. et al. Ground and excited state properties of new porphyrin based dyads: a combined theoretical and experimental study. J. Phys. Chem. A 116, 10736–10744 (2012).
-
Kantorovich, L. V. Mathematical methods of organizing and planning production. Manag. Sci. 6, 366–422 (1960).
-
Ramdas, A., García Trillos, N. & Cuturi, M. On Wasserstein two-sample testing and related families of nonparametric tests. Entropy 19, 47 (2017).
-
Kotlobay, A. A. et al. Genetically encodable bioluminescent system from fungi. Proc. Natl Acad. Sci. USA 115, 12728–12732 (2018).
-
Gallo, G., Longo, G., Pallottino, S. & Nguyen, S. Directed hypergraphs and applications. Discret. Appl. Math. 42, 177–201 (1993).
-
Chen, G. E. et al. Complete enzyme set for chlorophyll biosynthesis in Escherichia coli. Sci. Adv. 4, eaaq1407 (2018).
-
Bioucas-Dias, J. M. et al. Hyperspectral unmixing overview: geometrical, statistical, and sparse regression-based approaches. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 5, 354–379 (2012).
-
Xu, L., Li, J., Wong, A. & Peng, J. K-P-Means: a clustering algorithm of K ‘purified’ means for hyperspectral endmember estimation. IEEE Geosci. Remote Sens. Lett. 11, 1787–1791 (2014).
-
Prades, J., Safont, G., Salazar, A. & Vergara, L. Estimation of the number of endmembers in hyperspectral images using agglomerative clustering. Remote Sensing 12, 3585 (2020).
-
Wegele, R., Tasler, R., Zeng, Y., Rivera, M. & Frankenberg-Dinkel, N. The heme oxygenase (s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279, 45791–45802 (2004).
-
Maiti, A. et al. Structural and photophysical characterization of the small ultra-red fluorescent protein. Nat. Commun. 14, 4155 (2023).
-
Boo, A. et al. Synthetic microbe-to-plant communication channels. Nat. Commun. 15, 1817 (2024).
-
Chen, G. E. & Hunter, C. N. Engineering chlorophyll, bacteriochlorophyll, and carotenoid biosynthetic pathways in Escherichia coli. ACS Synth. Biol. 12, 2236–2244 (2023).
-
Saga, Y. et al. Selective oxidation of B800 bacteriochlorophyll a in photosynthetic light-harvesting protein LH2. Sci. Rep. 9, 3636 (2019).
-
Becker, M., Nagarajan, V. & Parson, W. W. Properties of the excited singlet states of bacteriochlorophyll a and bacteriopheophytin a in polar solvents. J. Am. Chem. Soc. 113, 6840–6848 (1991).
-
Yokobayashi, Y., Weiss, R. & Arnold, F. H. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA 99, 16587–16591 (2002).
-
Tabor, J. J., Levskaya, A. & Voigt, C. A. Multichromatic control of gene expression in Escherichia coli. J. Mol. Biol. 405, 315–324 (2011).
-
Fernandez-Rodriguez, J., Moser, F., Song, M. & Voigt, C. A. Engineering RGB color vision into Escherichia coli. Nat. Chem. Biol. 13, 706–708 (2017).
-
Bradley, A. P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 30, 1145–1159 (1997).
-
Gouterman, M. Study of the effects of substitution on the absorption spectra of porphin. J. Chem. Phys. 30, 1139–1161 (1959).
-
Taniguchi, M., Bocian, D. F., Holten, D. & Lindsey, J. S. Beyond green with synthetic chlorophylls—connecting structural features with spectral properties. J. Photoch. Photobio. C 52, 100513 (2022).
-
Gouterman, M. Spectra of porphyrins. J. Mol. Spectrosc. 6, 138–163 (1961).
-
Taniguchi, M. & Lindsey, J. S. Absorption and fluorescence spectral database of chlorophylls and analogues. Photochem. Photobiol. 97, 136–165 (2021).
-
Lakomy, I. et al. C45– and C50-carotenoids. Part 8. Synthesis of (all-E,2S,2′S)-bacterioruberin, (all-E,2S,2′S)-monoanhydrobacterioruberin, (all-E,2S,2′S)-bisanhydrobacterioruberin, (all-E,2R,2′R)-3,4,3′,4′-tetrahydrobisanhydrobacterioruberin, and (all-E,S)-2-isopentenyl-3,4-dehydrorhodopin. Helvetica 80, 472–486 (2010).
-
Levin, I., Liu, M., Voigt, C. A. & Coley, C. W. Merging enzymatic and synthetic chemistry with computational synthesis planning. Nat. Commun. 13, 7747 (2022).
-
Sankaranarayanan, K. et al. Similarity based enzymatic retrosynthesis. Chem. Sci. 13, 6039–6053 (2022).
-
Probst, D. et al. Biocatalysed synthesis planning using data-driven learning. Nat. Commun. 13, 964 (2022).
-
Koch, M., Duigou, T. & Faulon, J.-L. Reinforcement learning for bioretrosynthesis. ACS Synth. Biol. 9, 157–168 (2019).
-
Lin, G.-M., Warden-Rothman, R. & Voigt, C. A. Retrosynthetic design of metabolic pathways to chemicals not found in nature. Curr. Opin. Syst. Biol. 14, 82–107 (2019).
-
Koscher, B. A. et al. Autonomous, multiproperty-driven molecular discovery: from predictions to measurements and back. Science 382, eadi1407 (2023).
-
Gómez-Bombarelli, R. et al. Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach. Nat. Mater. 15, 1120–1127 (2016).
-
Chemla, Y., Sweeney, C. J., Wozniak, C. A. & Voigt, C. A. Design and regulation of engineered bacteria for environmental release. Nat. Microbiol. 10, 281–300 (2025).
-
Bloch, S. E. et al. Biological nitrogen fixation in maize: optimizing nitrogenase expression in a root-associated diazotroph. J. Exp. Bot. 71, 4591–4603 (2020).
-
Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988).
-
Riniker, S. & Landrum, G. A. Better informed distance geometry: using what we know to improve conformation generation. J. Chem. Inf. Model. 55, 2562–2574 (2015).
-
Halgren, T. A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 17, 490–519 (1996).
-
Bannwarth, C., Ehlert, S. & Grimme, S. GFN2-xTB—an accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J. Chem. Theory Comput. 15, 1652–1671 (2019).
-
Cossi, M. & Barone, V. Time-dependent density functional theory for molecules in liquid solutions. J. Chem. Phys. 115, 4708–4717 (2001).
-
Chemla, Y. et al. Parallel engineering of environmental bacteria and performance over years under jungle-simulated conditions. PLoS ONE 17, e0278471 (2022).
-
Johnson, S. C. Hierarchical clustering schemes. Psychometrika 32, 241–254 (1967).
-
Jiang, J., Liu, D., Gu, J. & Süsstrunk, S. What is the space of spectral sensitivity functions for digital color cameras? In Proc. 2013 IEEE Workshop on Applications of Computer Vision (WACV) 168–179 (IEEE, 2013).
-
Chemla, Y. Hyperspectral reporters for long-distance and wide-area detection of gene expression in living bacteria. Zenodo https://doi.org/10.5281/zenodo.14756888 (2025).
-
Levin, I. et al. VoigtLab/bioHSI: v.1.0.0. Zenodo https://doi.org/10.5281/zenodo.14827800 (2025).
-
Levin, I. et al. VoigtLab/npspec-webapp: v1.0.0. Zenodo https://zenodo.org/records/14827805 (2025).